https://sourceforge.net/projects/arcadiapathways/
Follow the tour guide. The SBML file used in the screenshots can be downloaded here.
For more SBML files, you can visit Biomodels.net.
Please refer to:
As the amount of data available on biological systems
increases, so does the need for computing
tools supporting their analysis.
In
particular, by enabling the interactive exploration on various kind
of pathways, visualisation software provides considerable
assistance in making
sense of complex networks.
Unfortunately, existing tools struggle to address adequately the
specific case of metabolic
pathways: automatic layouts computed by generic
graph-drawing algorithms usually require time-consuming manual
adjustments.
Arcadia has been designed specifically to provide relevant visualisation options for metabolic pathways.
Arcadia is a viewer, not an editor: this means a simpler interface, offering multiple perspectives on the same data, with a focus on navigation.
As a light-weight, standalone component, Arcadia is easy to deploy and maintain. In order to ensure interoperability with other tools, an effort is made to support existing or emerging standards such as SBML and SBGN.
A reference framework for the development of Arcadia is Utopia: an open-source, interoperable set of desktop tools for protein analysis, built according to the Model/View/Controller pattern. This approach, along with Object Oriented Programming, makes up for flexible, evolutive software.
Written in C++, Arcadia makes use of powerful existing open-source libraries:
Cross-platform, the code can be compiled to run on Mac, UNIX or Windows systems.
Arcadia enables navigation between multiple interconnected views of the same model:
Metadata annotations identifying different types of species and reactions are used to define their visual appearance, in accordance with the SBGN recommendations.
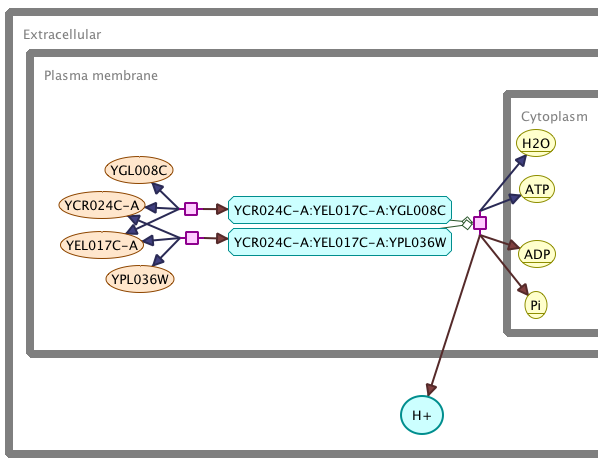
Intuitive, context-sensitive mouse-controls can be used to apply local layout strategies to particular subsets of the network. Default behaviours have been defined for typical domain-specific concepts (e.g. "modifier", "cofactor", etc.) resulting in a semi-automated layout.
The prototype has been successfully tested on a dozen of
small-scale models (< 500 nodes = species + reactions).
The layouts obtained after a few quick and easy steps are very similar
to the hand-drawn diagram used as a reference.
The source code of the current version is available on sourceforge.net
driven by semantic information extracted from: